


Our groups interest lies in understanding the basis of biological heterogeneity using computational approaches applied to ‘omics data in a disease context. We employ artificial intelligence (AI) and data science (DS) methods, not only to address the underlying causes of this heterogeneity, but also the interactions between different molecular ‘omics layers. We have a strong specialisation in the application area of oncology, but are also very interested in immunology and neurodegenerative diseases.
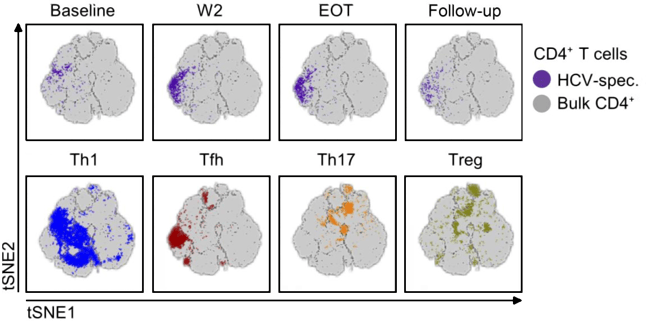
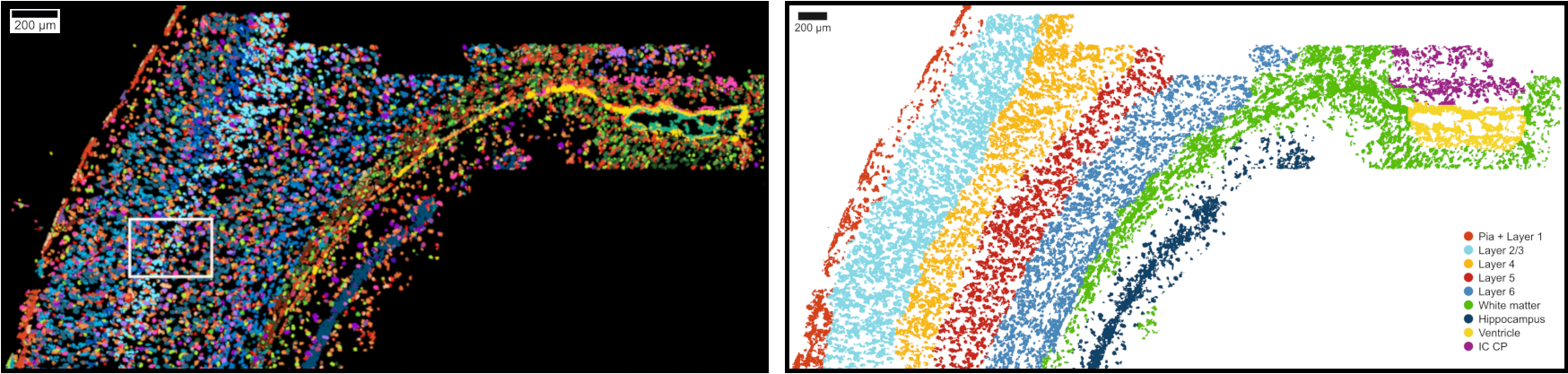
We believe that by better understanding global and local patterns in biological data, we can better differentiate the mechanistics behind biological and molecular processes from technical noise. As such, we rely on establishing close ties with experimental collaborators who are able to provide us with large scale and high dimensional datasets profiled using state of the art and exotic profiling techniques. We aim to take advantage of recent technological advances that are able to resolve single cell, spatial, temporal and genetic heterogeneity.
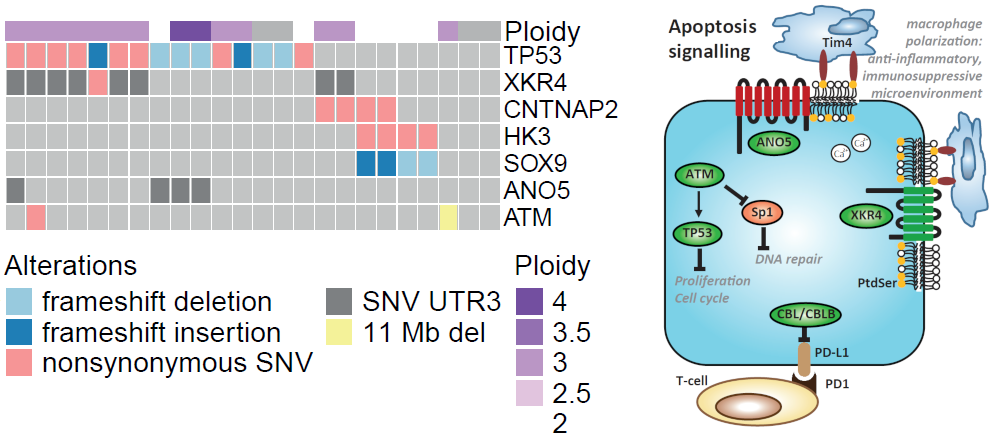
The group has a strong core expertise in analysis and interpretation of cancer multiomics data, with exposure to a wide number of omics layers and tumour entities. We are in particular very interested in understanding the heterogeneous patient response to drugs and are interested in accurate classification of tumour entities, patient specific molecular drivers, tumour microenvironment and TILs, tumour evolution and heterogeneity, and germline predisposing factors.

Naveed Ishaque, Ph.D.
Group leader Computational Oncology
The headline and subheader tells us what you're offering, and the form header closes the deal. Over here you can explain why your offer is so great it's worth filling out a form for.
Remember:
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor

Our colleagues from Northestern University in Boston are organising the 18th INTERNATIONAL CONFERENCE ON SYSTEMS BIOLOGY OF HUMAN DISEASE – SBHD 2026 in Boston from June 08-10. This years Conference

The Berlin Institute of Health at Charité (BIH) coordinates the pan-European research initiative "EU-CiP" (European Cancer Information Portal). Supported by 12 million euros from the EU’s Horizon

The Digital Health Center once again organises the INTERNATIONAL CONFERENCE ON SYSTEMS BIOLOGY OF HUMAN DISEASE – SBHD 2025 in Berlin from June 16-18. Don’t miss this opportunity to participate in a

Our colleagues at Vanderbilt University organise the 16th INTERNATIONAL CONFERENCE ON SYSTEMS BIOLOGY OF HUMAN DISEASE – SBHD 2024 this year from June 10-12. Don’t miss the opportunity to participate
The Hub for Innovations in Digital Health (HiDiH) brings together two independent sites of excellent research and development in Berlin and in Heidelberg. HIDIH’s major branch in Berlin is the Center for Digital Health at the Charité and the Berlin Institute of Health (BIH).
If we caught your attention, you are interested in our work and would like to get in touch with us, please contact us via franziska.mueller@bih-charite.de



