

The Synthetic Biology group focuses on the development of tools for precision perturbation and control of cellular processes in cells and animals. Our main research branches are:
We are embedded into a great network of collaboration partners in Heidelberg (Prof. Dr. Dirk Grimm, Dr. Kathleen Börner) and have multiple, ongoing national and international collaborations, e.g. with Bruno E. Correia at the EPFL, Lausanne, and Prof. Dr. Pam Silver at Harvard Medical School.
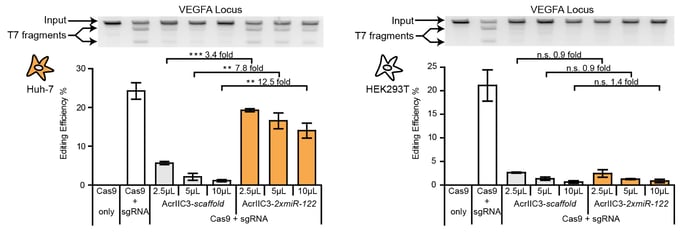
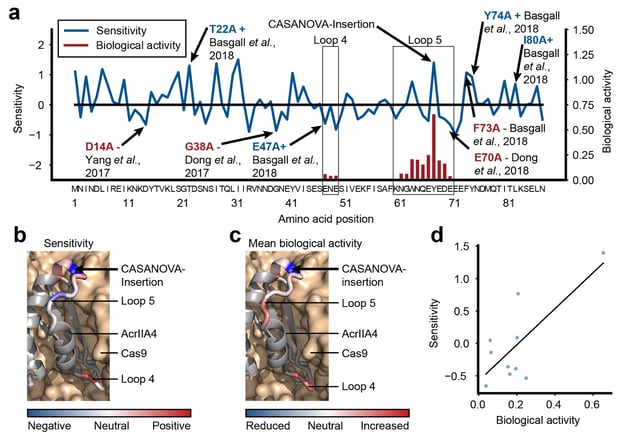
Anti-CRISPR proteins are potent inhibitors of CRISPR effectors. We engineer light-switchable derivatives of natural anti-CRISPRs by embedding photosensor domains, most typically the light-oxygen-voltage 2 (LOV2) domain from Avena sativa, into Acr structures (Bubeck & Hoffmann et al., 2019; Mathony and Hoffmann et al., submitted). The resulting, chimeric inhibitors block Cas9 in the dark, but release its activity upon blue light irradiation. This approach which we named CASANOVA, for CRISPR-Cas activity switching by novel, optogenetic variants of anti-CRISPR proteins, facilitates light-dependent genome editing and epigenome editing and is used in our lab to study genome regulatory processes. While CASANOVA was initially limited to the S. pyogenes Cas9, we are currently extending this original strategy to other Cas9 orthologues and Cas12. We are also developing designer Acrs that outperform their natural counterparts with respect to inhibition potency (Mathony, Harteveld and Schmelas et al., bioRxiv, 2019).
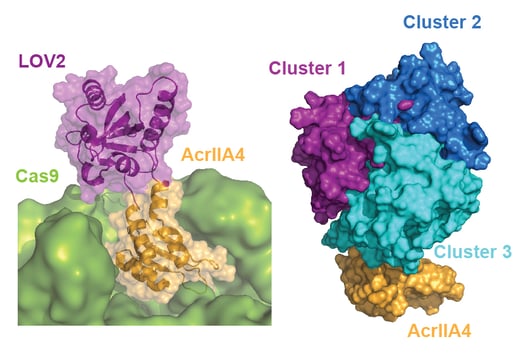
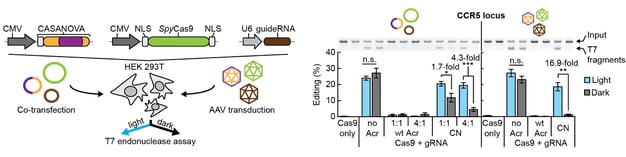
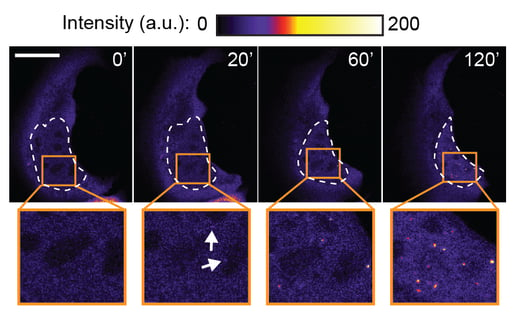
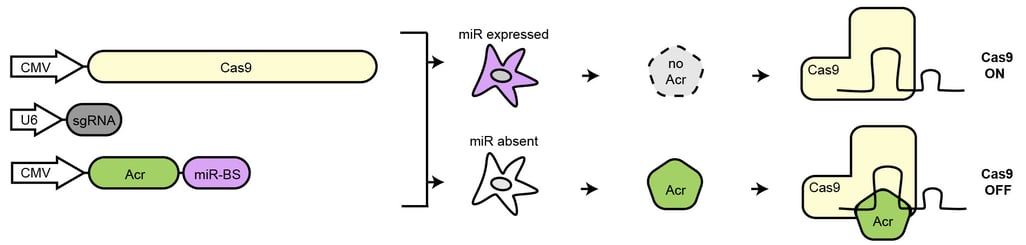
One of our central aims is to support the clinical translation of the CRISPR technologies by developing safe and efficient AAV-CRISPR vectors (Senis et al., 2014). Towards this goal, we closely collaborate with Prof. Dr. Dirk Grimm at Heidelberg University clinics. We use synthetic biology approaches to create switches and circuits that confine Cas9 activity to selected cell types, e.g. by coupling Cas9 activity to the abundance of cell type-specific microRNAs (Hoffmann et al., 2019). We also combine wet lab experiments with mathematical modeling to fine-tune Cas9 activity to desired levels, thereby enabling the kinetic insulation of ON- and OFF-target editing events (Aschenbrenner & Kallenberger et al., 2020).
By training neural networks on thousands of protein sequences and corresponding 3D protein structures, we aim at creating algorithms that can inform protein design (Upmeier zu Belzen et al., 2019). Applications range from the prediction of engineering hotspots to facilitate the development of switchable proteins to the re-design of enzymes and creation of immune “stealth” protein therapeutics.

Dominik obtained a M.Sc. in Molecular Biotechnology at Heidelberg University. In 2016, he completed his PhD at the German Cancer Research Center and took over the Synthetic Biology group.
Dr. Dominik Niopek
Group leader at BioQuant-Zentrum

Aschenbrenner, S.*, Kallenberger, S.*, Hoffmann, M. D., Huck, A., Eils, R.§ & Niopek, D.§ (2020). Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Science Advances, 6(6), doi:10.1126/sciadv.aay0187
Hoffmann, M. D., Aschenbrenner, S., Grosse, S., Rapti, K., Domenger, C., Mastel, M., Eils, R.§, Grimm, D.§ & Niopek, D.§ (2019). Cell-specific CRISPR-Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Research, 47(13), doi:10.1093/nar/gkz271
Upmeier zu Belzen, J., Bürgel, T., Holderbach, S., Bubeck, F., Adam, L., Gandor, C., Klein, M., Mathony, J., Pfuderer, P. L., Platz, L., Przybilla, M., Schwendemann, M., Heid, D., Hoffmann, M. D., Jendrusch, M., Schmelas, C., Waldhauer, M., Lehmann, I., Niopek, D.§ & Eils, R.§ (2019). Leveraging implicit knowledge in neural networks for functional dissection and engineering of proteins. Nature Machine Intelligence 1, 225-235. doi: 10.1038/s42256-019-0049-9
Bubeck, F., Hoffmann, M. D., Harteveld, Z., Aschenbrenner, S., Bietz, A., Waldhauer, M. C., Börner, K., Fakhiri, J., Schmelas, C., Dietz, L., Grimm, D., Correia, B. E., Eils, R.§ & Niopek, D.§ (2018). Engineered anti-CRISPR proteins for optogenetic control of CRISPR/Cas9. Nature Methods. 15(11), 924-927, doi:10.1038/s41592-018-0178-9
Niopek, D., Wehler, P., Roensch, J., Eils, R.§ & Di Ventura, B.§ (2016). Optogenetic Control of Nuclear Protein Export. Nature Communication. 7. doi: 10.1038/ncomms10624
*these authors contributed equally
§corresponding authorThe headline and subheader tells us what you're offering, and the form header closes the deal. Over here you can explain why your offer is so great it's worth filling out a form for.
Remember:
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor
Feature Title
Lorem ipsum dolor sit amet
Consectetur adipiscing elit
Integer ornare lectus ut arcu
Tincidunt porttitor

The Digital Health Center once again organises the INTERNATIONAL CONFERENCE ON SYSTEMS BIOLOGY OF HUMAN DISEASE – SBHD 2025 in Berlin from June 16-18. Don’t miss this opportunity to participate in a

Our colleagues at Vanderbilt University organise the 16th INTERNATIONAL CONFERENCE ON SYSTEMS BIOLOGY OF HUMAN DISEASE – SBHD 2024 this year from June 10-12. Don’t miss the opportunity to participate

Am 11.10.2023 wurde Prof. Roland Eils im Tagesspiegel als einer der 100 wichtigsten Köpfe der Hauptstadt-Wissenschaft gewürdigt. So schreibt der Tagesspiegel: "Um Big Data dreht sich alles in der

The Lindau Nobel Laureate Meetings are annual conferences where some of the brightest minds in science converge to exchange knowledge, foster collaboration, and inspire the next generation of
The Hub for Innovations in Digital Health (HiDiH) brings together two independent sites of excellent research and development in Berlin and in Heidelberg. HIDIH’s major branch in Berlin is the Center for Digital Health at the Charité and the Berlin Institute of Health (BIH).
If we caught your attention, you are interested in our work and would like to get in touch with us, please contact us via franziska.mueller@bih-charite.de



